Genotyping, sequencing, and profiling
Structure of DNA
Deoxyribonucleic acid is a large macromolecule composed of antiparallel (going in opposite directions) chains of nucleotides. Each nucleotide consists of a pair of nucleobases (Adenine paired with Thymine, Cytosine paired with Guanine).
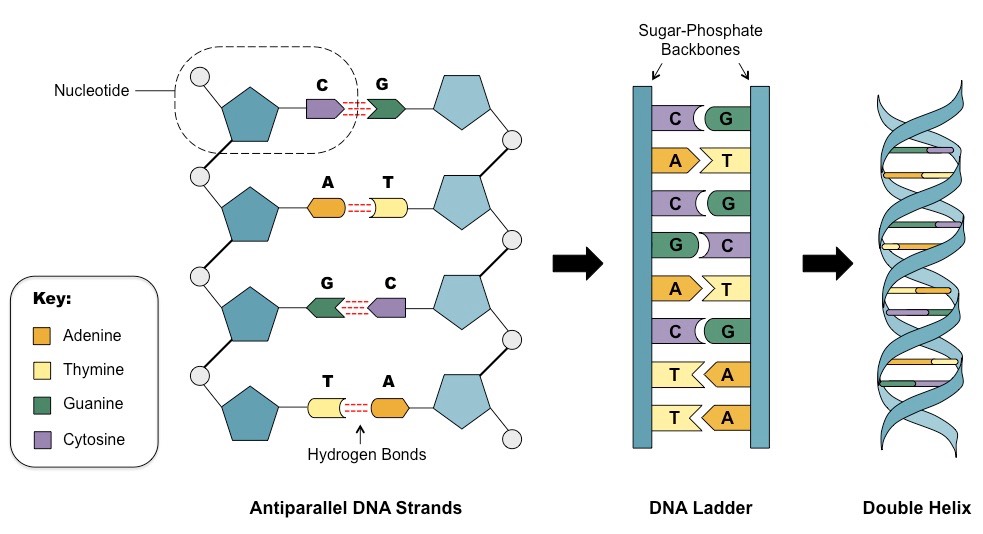
Each DNA molecule is packed into a chromosome. Humans are diploid, with two versions of each chromosome inherited from each parent, and have 22 pairs of autosomes and 1 pair of sex chromosomes). The human genome contains around 3 billion basepairs.
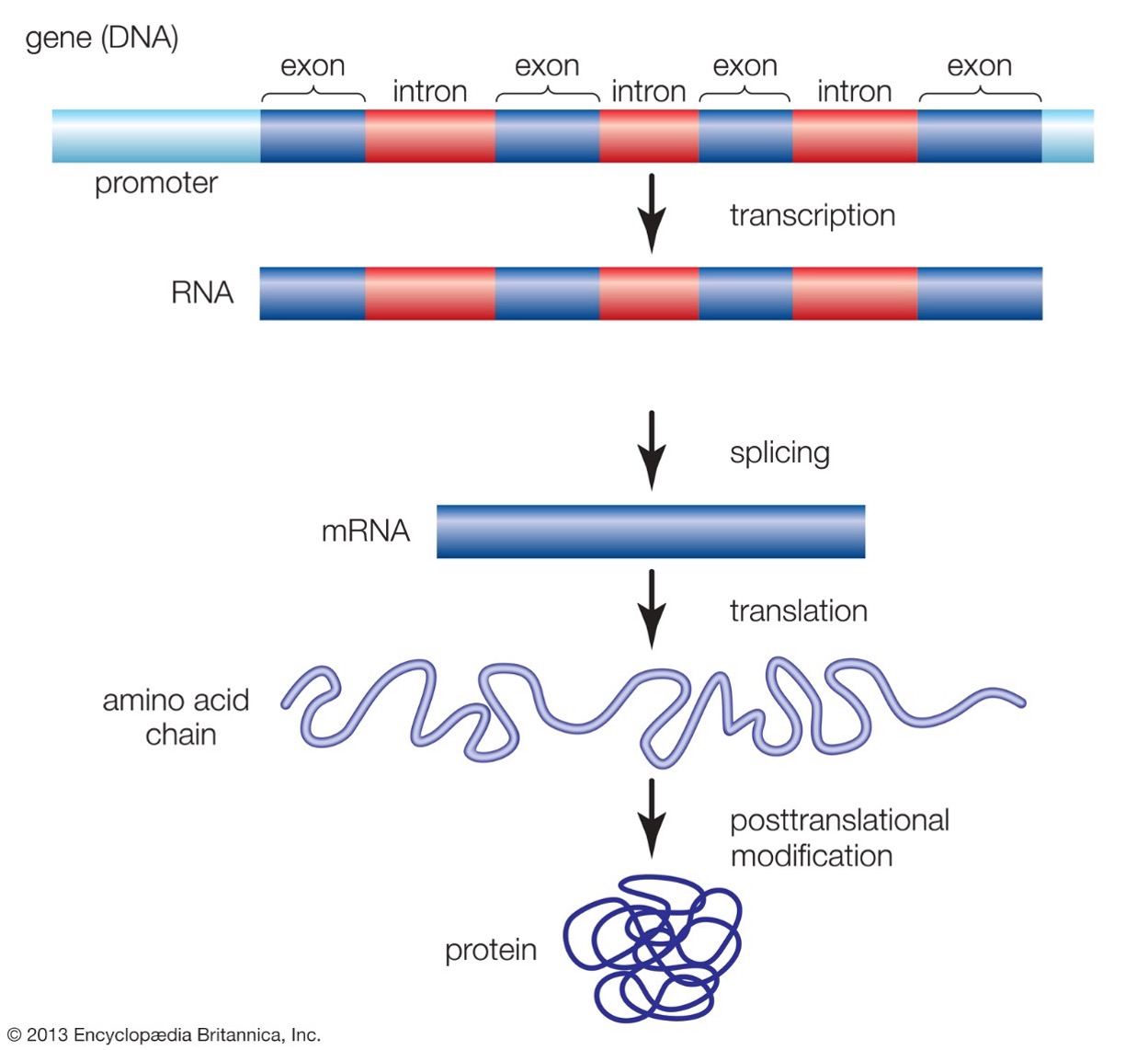
A portion of the genome contains genes that encode proteins. A gene has a promotor region, which is a DNA sequence that where and how much the gene is expressed. A gene contains a number of exons (coding sequence, determining the amino acide composition of the protein) and introns (non-coding sequence). DNA is transcripted into RNA, which is then spliced into mRNA and then becomes the template for the animo acid chain which folds into a protein.
Microarrays
Modern DNA genotyping is performed with microarrays. Microarrays contain thousands of labelled probes of short DNA sequences of different genotypes. The target DNA hybridises with the matching probe. Probes are coupled to a dye that is read with a laser. The intensity of the alternative probes is used to cluster each sample to determine the genotype at each locus on the array.
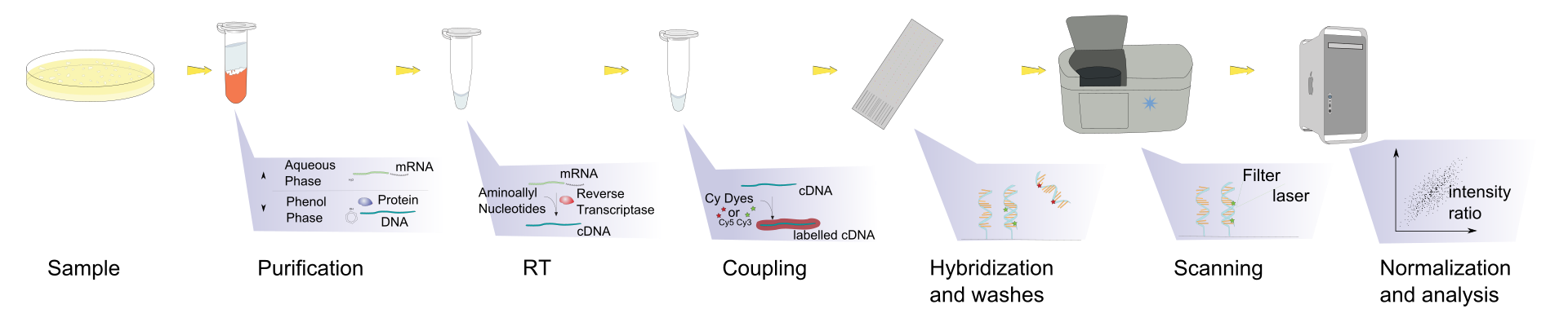
Watch a Microarray Method for Genetic Testing Video here
Because microarray genotypes only assay a small portion of the genome, imputation is often used to statistically fill in the non-genotyped portions of the genome. Imputation compares the genotyped target sample to the full genome sequence of a reference dataset. This is effective for variants that are common in the population.
Sequencing
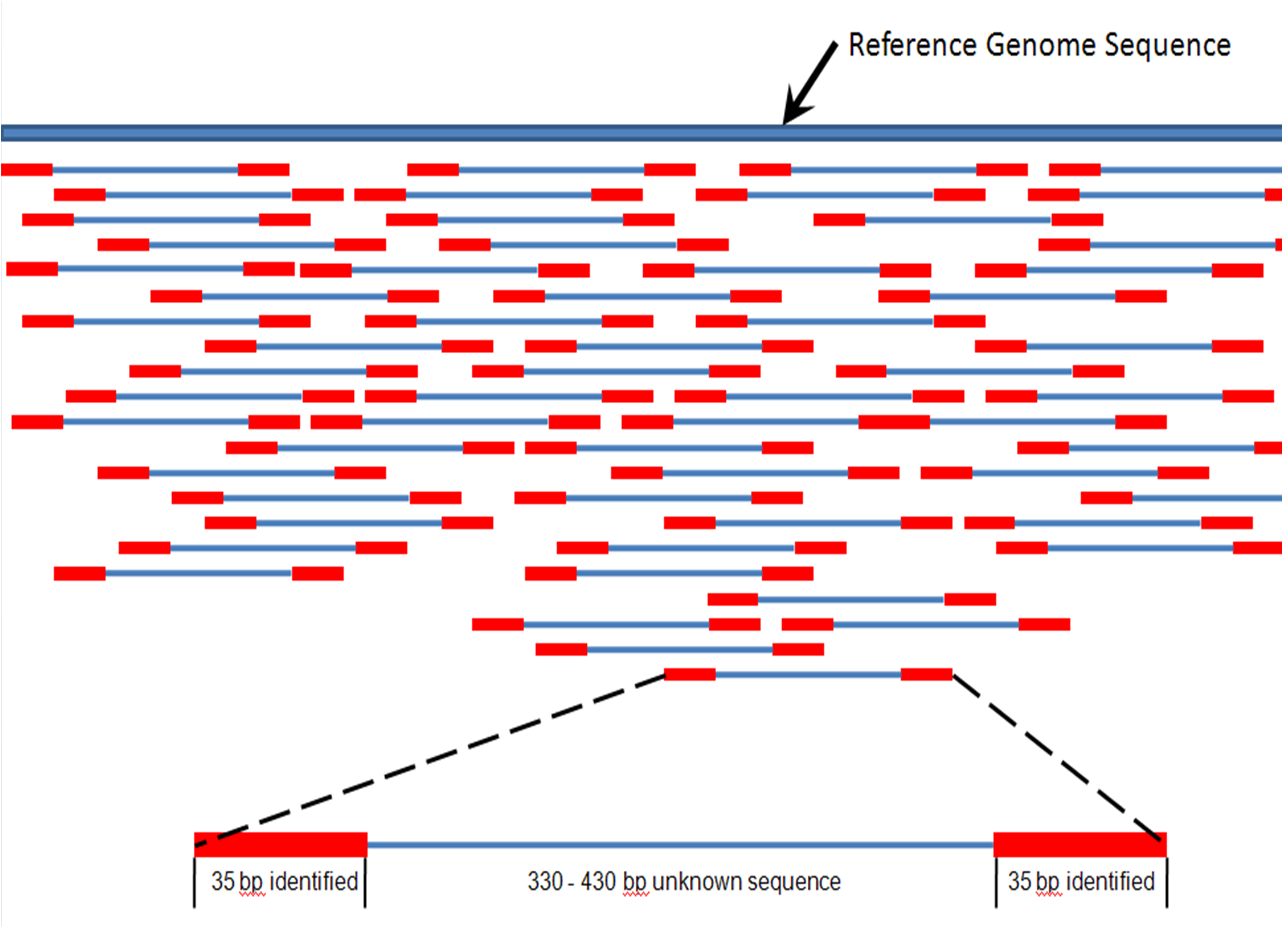
In contrast to genotyping, which only assays around 1 million (< 0.1%) parts of the genome, sequencing captures 10x (for exome sequencing) or 1000x (for whole genome sequencing) the amount of genomic information. While genotyping can only assay genetic variants that have previously been identified, sequencing can assay previously unknown genetic variants. Sequencing is typically peformed by cutting DNA into small fragments, reading each fragment (using different technologies), and then reassembling the sequences using a reference genome.
Exome sequencing captures the exomes of genes (that code proteins) while whole genome sequencing captures most of the genome (usually excluding segements that are difficult to sequence such as repetitive elements or portions of the genome that are near the telomeres or centromeres).
Genomic coordinates
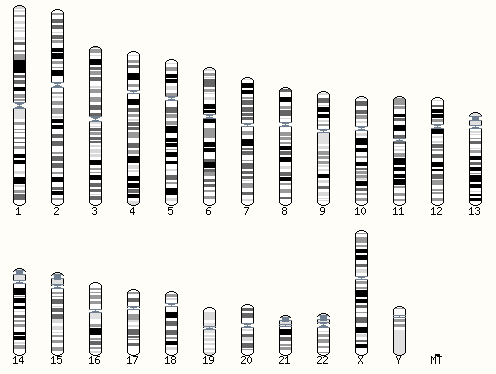
Every portion of the genome is assigned a standard coordinate position. Chromosomes were numbered based on their apparent size based on traditional light microscopy. The autosomes are numbered 1-22 and the allosomes (X = 23, Y = 24, MT = 25). Within each chromosome, basepair positions are counted from one end of the chromosome to the other. As more is learned about the structure of the genome, the coordinate system gets periodically updated as a “build” of the genome. Most datasets currently available use a build called “GRCh37” (Genome Reference Consortium Human Build 37) or “hg19” (Human Genome Issue HG-19). More recent datasets have moved to a new build, called “GRCh38/hg38“.